












RNA tertiary motifs play an important role in RNA folding. To understand the complex organization of RNA tertiary interactions, we compiled a dataset containing 54 high-resolution RNA crystal structures. Seven RNA tertiary motifs (coaxial helix, A-minor, ribose zipper, pseudoknot, loop-loop receptor, tRNA D-loop;T-loop, and kissing hairpin) were searched by different computer programs. For the non-redundant RNA dataset, 613 RNA tertiary interactions were found. Most of these 3D interactions occur in the 16S and 23S rRNAs. Exhaustive search of these motifs revealed diversity of A-minor interactions, and other loop-loop receptor interactions similar to the tetraloop-tetraloop receptor. Correlation between motifs, such as pseudoknot or coaxial helix with A-minor, shows that they can form composite motifs. These findings may lead to tertiary structure constraints for RNA 3D prediction.
Display annotation diagram by molecular feature:
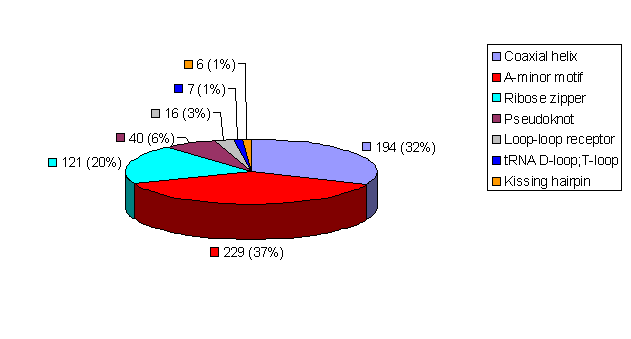
For the non-redundant 54-structure dataset, 613 RNA tertiary interactions were found. The seven motifs are unevenly distributed in RNA structures. Coaxial helices, A-minor interactions, and ribose zippers account for 89% of the total tertiary motifs. The complete list of teritary motif distribution in the non-redundant dataset is here.
 |
Coaxial helix | |
 |
 |
A-minor motif |
 |
 |
Ribose zipper |
 |
 |
Pseudoknot |
 |
 |
Kissing hairpin |
 |
 |
tRNA D-loop;T-loop |
 |
 |
Loop-loop receptor |