Dual Topology: 4_14
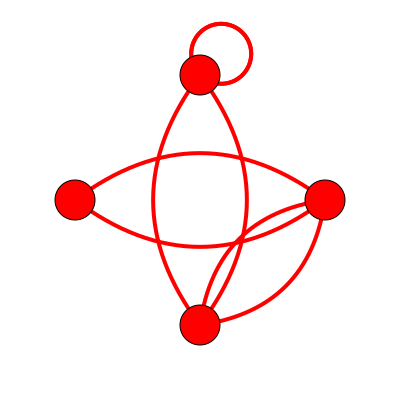
Topological Aspects
- 4 Vertices
- Eigenvalues
- 0.0000000000
- 1.1715729237
- 4.0000000000
- 6.8284273148
Examples of actual RNAs corresponding to this structure from RNA databases: 37
| PDB_Chain | Type | Discovery Method |
|---|---|---|
| 1KXK_A | ai5g group II Self-splicing intron | X-RAY DIFFRACTION |
| 1P5O_A | 77-MER | SOLUTION NMR |
| 1P5P_A | 77-MER | SOLUTION NMR |
| 1XJR_A | s2m RNA | X-RAY DIFFRACTION |
| 3U4M_B | RNA (80-MER) | X-RAY DIFFRACTION |
| 3U56_B | RNA (80-MER) | X-RAY DIFFRACTION |
| 3UMY_B | RNA (80-MER) | X-RAY DIFFRACTION |
| 4ERD_CD | 5'-R(P*GP*GP*UP*CP*GP*AP*CP*AP*UP*CP*UP*UP*CP*GP*G | X-RAY DIFFRACTION |
| 4KZD_R | RNA (84-MER) | X-RAY DIFFRACTION |
| 4KZE_R | RNA (84-MER) | X-RAY DIFFRACTION |
| 4N0T_B | U6 SNRNA | X-RAY DIFFRACTION |
| 4Q9Q_R | SPINACH RNA APTAMER | X-RAY DIFFRACTION |
| 4Q9R_R | SPINACH RNA APTAMER | X-RAY DIFFRACTION |
| 4QG3_B | FRAGMENT OF 23S RRNA | X-RAY DIFFRACTION |
| 4QVI_B | fragment of 23S rRNA | X-RAY DIFFRACTION |
| 5IEM_A | RNA (57-MER) | SOLUTION NMR |
| 5NPM_B | 23S RIBOSOMAL RNA | X-RAY DIFFRACTION |
| 5UNE_AB | RNA (47-MER) | X-RAY DIFFRACTION |
| 6B14_R | RNA (86-MER) | X-RAY DIFFRACTION |
| 6B3K_R | RNA (83-MER),RNA (83-MER) | X-RAY DIFFRACTION |
| 1D4R_BA | 29-MER OF MODIFIED SRP RNA HELIX 6 | X-RAY DIFFRACTION |
| 2J28_8 | 4.5S SIGNAL RECOGNITION PARTICLE RNA | ELECTRON MICROSCOPY |
| 3J45_5 | 23S RIBOSOMAL RNA | ELECTRON MICROSCOPY |
| 3J45_4 | 23S ribosomal RNA | ELECTRON MICROSCOPY |
| 3JB9_C | U5 snRNA | ELECTRON MICROSCOPY |
| 3RW6_H | constitutive transport element(CTE)of Mason-Pfizer | X-RAY DIFFRACTION |
| 3SIV_CF | U4atac snRNA | X-RAY DIFFRACTION |
| 4MGN_A | glyQS T box riboswitch | X-RAY DIFFRACTION |
| 4V61_BC | 4.8S rRNA | ELECTRON MICROSCOPY |
| 4V8Y_CN | EUKARYOTIC RIBOSOMAL L1_RRNA | ELECTRON MICROSCOPY |
| 5AKA_7 | 4.5S ribosomal RNA | ELECTRON MICROSCOPY |
| 5DCV_D | RNA (47-MER) | X-RAY DIFFRACTION |
| 5LI0_B | 5S ribosomal RNA | ELECTRON MICROSCOPY |
| 5LYS_B | 7SK RNA | X-RAY DIFFRACTION |
| 5LYU_B | 7SK RNA | X-RAY DIFFRACTION |
| 5LYV_B | RNA (57-MER) | X-RAY DIFFRACTION |
| 5XTM_D | RNA (47-MER) | X-RAY DIFFRACTION |