Dual Topology: 7_2019
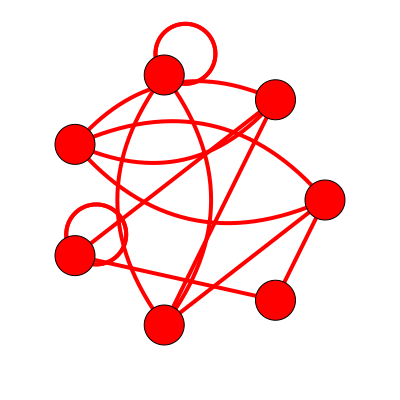
Topological Aspects
- 7 Vertices
- Eigenvalues
- 0.0000000000
- 0.8503010869
- 1.7848318815
- 2.3819661140
- 4.6180338860
- 5.0321302414
- 7.3327369690
Examples of actual RNAs corresponding to this structure from RNA databases: 24
| PDB_Chain | Type | Discovery Method |
|---|---|---|
| 2GIS_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 2YDH_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 2YGH_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3GX2_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3GX3_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3GX5_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3GX7_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3IQN_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3IQR_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 4AOB_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 4B5R_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FJC_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK1_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK2_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK3_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK4_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK5_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK6_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKD_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKE_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKF_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKG_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKH_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3V7E_C | SAM-I riboswitch aptamer with an engineered helix | X-RAY DIFFRACTION |