Tree Topology: 6_5
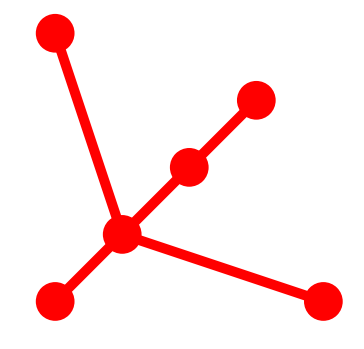
Topological Aspects
- 6 Vertices
- Eigenvalues 0.0000000000
0.4858630598
1.0000000000
1.0000000000
2.4280066490
5.0861301422
Examples of actual RNAs corresponding to this structure from RNA databases: 32
| PDB_Chain | Type | Discovery Method |
|---|---|---|
| 2NOQ_A | CrPV IRES | ELECTRON MICROSCOPY |
| 2YGH_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3G8S_Q | GlmS ribozyme | X-RAY DIFFRACTION |
| 3G8T_R | glmS glucosamine-6-phosphate activated ribozyme | X-RAY DIFFRACTION |
| 3G96_R | GlmS ribozyme | X-RAY DIFFRACTION |
| 3G9C_S | GlmS ribozyme | X-RAY DIFFRACTION |
| 3GX6_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3GX7_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3HHN_E | Class I ligase ribozyme, self-ligation product | X-RAY DIFFRACTION |
| 3IQP_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3IVK_M | class I ligase product | X-RAY DIFFRACTION |
| 3L3C_Q | GlmS ribozyme | X-RAY DIFFRACTION |
| 3Q1Q_C | TRNA (PHE) | X-RAY DIFFRACTION |
| 3Q1R_C | TRNA (PHE) | X-RAY DIFFRACTION |
| 3R1H_F | CLASS I LIGASE RIBOZYME | X-RAY DIFFRACTION |
| 3R1L_C | CLASS I LIGASE RIBOZYME | X-RAY DIFFRACTION |
| 4D5N_X | CRICKET PARALYSIS VIRUS IRES RNA | ELECTRON MICROSCOPY |
| 4D61_j | CRICKET PARALYSIS VIRUS IRES RNA | ELECTRON MICROSCOPY |
| 4FRG_B | cobalamin riboswitch aptamer domain | X-RAY DIFFRACTION |
| 4LCK_B | TRNA-GLY | X-RAY DIFFRACTION |
| 4OO8_B | RNA (97-MER) | X-RAY DIFFRACTION |
| 4RUM_A | NiCo riboswitch RNA | X-RAY DIFFRACTION |
| 4TZP_E | ENGINEERED TRNA | X-RAY DIFFRACTION |
| 4TZV_B | ENGINEERED TRNA | X-RAY DIFFRACTION |
| 4TZZ_B | ENGINEERED TRNAGLY | X-RAY DIFFRACTION |
| 4WU1_3L | tRNA-Tyr | X-RAY DIFFRACTION |
| 5F9R_A | RNA (116-MER) | X-RAY DIFFRACTION |
| 5VZL_B | single guide RNA (116-MER) | ELECTRON MICROSCOPY |
| 5WE4_y | Phe-tRNA-Phe | ELECTRON MICROSCOPY |
| 5WE6_y | Phe-tRNA-Phe | ELECTRON MICROSCOPY |
| 5Y36_B | SINGLE-GUIDE RNA | ELECTRON MICROSCOPY |
| 6GAW_AV | P-site fMet-tRNAMet, mitochondrial | ELECTRON MICROSCOPY |