Tree Topology: 7_7
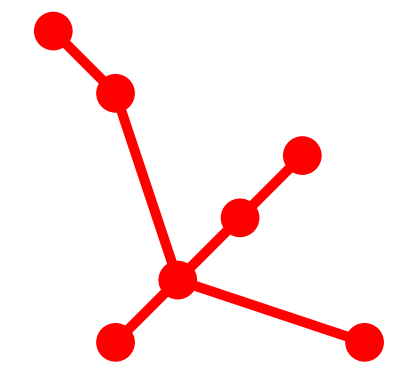
Topological Aspects
- 7 Vertices
- Eigenvalues 0.0000000000
0.3819660246
0.6086176038
1.0000000000
2.2271344662
2.6180338860
5.1642479897
Examples of actual RNAs corresponding to this structure from RNA databases: 28
| PDB_Chain | Type | Discovery Method |
|---|---|---|
| 2GIS_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 2YDH_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3GX2_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3GX3_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3GX5_A | RNA (94-MER) | X-RAY DIFFRACTION |
| 3IQN_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3IQR_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 3V7E_C | SAM-I riboswitch aptamer with an engineered helix | X-RAY DIFFRACTION |
| 4AOB_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 4B5R_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 4L81_A | SAM-I/IV variant riboswitch aptamer domain | X-RAY DIFFRACTION |
| 4Y1I_A | Lactococcus lactis yybP-ykoY riboswitch | X-RAY DIFFRACTION |
| 5FJC_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK1_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK2_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK3_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK4_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK5_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FK6_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKD_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKE_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKF_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKG_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5FKH_A | SAM-I RIBOSWITCH | X-RAY DIFFRACTION |
| 5IT7_4 | cricket paralysis virus IRES | ELECTRON MICROSCOPY |
| 5IT9_i | Cricket paralysis virus IRES RNA | ELECTRON MICROSCOPY |
| 6CB3_A | RNA (99-MER) | X-RAY DIFFRACTION |
| 6D9J_4 | CrPV IRES | ELECTRON MICROSCOPY |