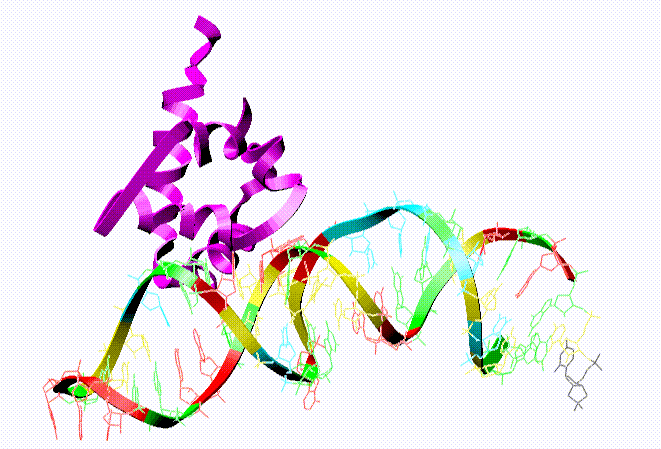RNA Structure
The RNA single-stranded polynucleotide chain folds in on itself in complex ways, forming hydrogen-bonded segments (e.g., A-U and G-C), imperfect with stems, loops, bulge loops, and junctions. The most stable fold for any particular RNA structure is the base paired structure that results in the minimum amount of free energy.
RNA folds form both secondary and tertiary structures. An RNA secondary structure shows how the bases pair to each other to produce stems, loops, and other secondary-structure motifs. The tertiary structure is considerably more complex; it is a representation of how the secondary structural elements interact with each other in three dimensional space. For example, a single strand RNA, that is identified by the Nucleic Acid Database as PR0021, has the following sequence of bases along its single strand:
RNA Sequence:
G G C U C U G U U U A C C A G G U C A G G U C C G A A A G G A A G C A G C C A A G G C A G A G C CCC
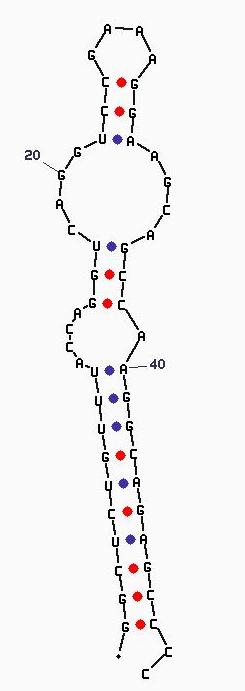
This sequence folds into the following secondary structure:
Image of PR0021 created using Zuker's mFold
|
Using the method described in the RNA Tree Graph Tutorial section of our website on how to create a secondary graph from a secondary structure, the secondary tree graph structure for this single strand RNA would appear as follows:
|
A pictoral view of the tertiary structure (complexed with a protein, purple) is provided in the Nucleic Acid Database, and it appears as follows: